Despite years of research, the genetic factors behind many human diseases and characteristics remain unknown. The inability to find the complete genetic causes of family traits such as height or the risk of type 2 diabetes has been called the "missing heritability" problem.
A new study by Princeton University researchers, however, suggests that missing heritability may not be missing after all — at least not in yeast cells, which the researchers used as a model for studying the problem. Published in the journal Nature, the results suggest that heritability in humans may be hidden due only to the limitations of modern research tools, but could be discovered if scientists know where (and how) to look.
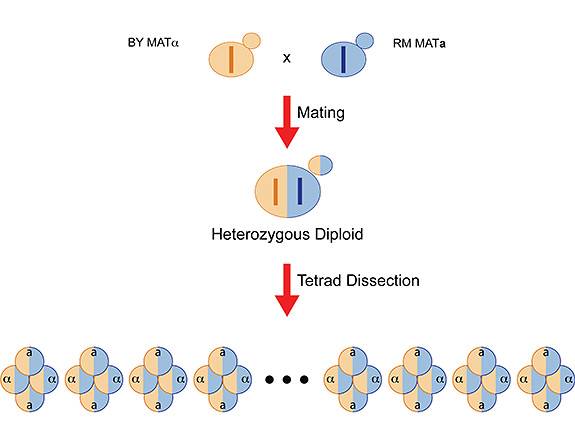
Researchers mated two yeast strains (BY from a laboratory and RM from a vineyard) and collected the offspring (tetrads) for genome-wide analysis to look for gene variations that contributed to survival. This experimental setup allowed the researchers to determine how much of the inherited survival traits could be detected by genome-wide scan and how much was "missing" or undetectable using a genome-wide scan. (Illustration by Joshua Bloom, Kruglyak Lab)
"The message of our study is that if you look hard enough you will find the missing heritability," said the senior researcher, Leonid Kruglyak, Princeton's William R. Harman '63 and Mary-Love Harman Professor in Genomics and a Howard Hughes Medical Institute Investigator. Kruglyak worked with first author Joshua Bloom, a Princeton graduate student; Wesley Loo, a 2010 Princeton graduate now a graduate student at Harvard University; Thuy-Lan Lite, Class of 2012, who is working at the National Institutes of Health for a year before starting graduate school; and Ian Ehrenreich, a past Princeton postdoctoral researcher now at the University of Southern California.
"We don't think there is some fundamental limitation — such as that there are things we don't understand about how genes behave — that is holding us back," Kruglyak said. "Instead, we should be able to detect the heritability in humans if we use the right tools."
Passed down from parent to child, genes determine not only eye color and other physical characteristics but also the risk of diseases. Some inherited diseases are caused by a mutation in a single gene. These single-gene disorders have well-defined patterns of inheritance that can be used to predict the chances that an individual will inherit the disease.
However, many diseases and physical traits arise due to multiple genes, multiple locations within genes, and even the regions of DNA between genes. Across the genome — which is an individual's total genetic content — small variations in DNA code can, when added together, increase or decrease the likelihood that a person will develop a disease or characteristic.
Height, for example, results from variations in DNA at multiple locations on the genome. Researchers have detected about 180 locations in the human genome where small alterations in the DNA code can have an influence on how tall or short a person is. Nonetheless, these locations account for only 13 percent of the expected contribution genetic code has on a person's height.
Type 2 diabetes also has missing heritability: About 40 identified genome locations are associated with the risk of developing the condition, but those account for only 10 percent of the estimated genetic influence. Finding the missing heritability for diseases like type 2 diabetes, Crohn's disease and schizophrenia could help inform prevention and treatment strategies.
In the present study, the researchers scanned the genomes of yeast cells for DNA variations — which can be thought of as spelling errors in the four-letter DNA code — and then matched those variations with qualities or characteristics inherited from the cells' parents. The researchers detected numerous DNA variations that, when added together, accounted for almost all of the offsprings' inherited characteristics, indicating that there was very little missing heritability in yeast.
Although the search for heritability was successful in yeast, finding missing heritability in humans is far more complicated, Kruglyak said. For example, interactions between genes can contribute to heritable traits, but such interactions are difficult to detect with genome-wide association studies (GWAS), which are the primary means by which geneticists look for DNA variations associated with diseases or traits. In addition, environmental factors such as nutrition also can influence gene activity, and these influences can be elusive to the genome-wide study. GWAS also may be inadequate at detecting common DNA spelling errors that have only small effects, or it may fail to find DNA variations that have a large effect but are rare.
The study sheds light on the role of nature (genetic factors) versus nurture (environmental factors) in determining traits and disease risk, according to Bert Vogelstein, director of the Ludwig Center at the Johns Hopkins University School of Medicine and a Howard Hughes Medical Institute Investigator.
"The nature versus nurture argument has been brewing for decades, both among scientists and the lay public, and 'missing heritability' has been problematic for the 'nature' component," said Vogelstein, who was not involved in the Princeton study.
"This beautiful study demonstrates that the genetic basis for heritability (nature) can be precisely defined if extensive, well-controlled experiments can be performed," Vogelstein said. "Though the results were obtained in a model organism, I would be surprised if they didn't apply, at least in part, to higher organisms, including humans."
Kruglyak said that one approach to finding the missing heritability in humans might be to apply genome-wide scans to large families, rather than focusing on large populations as is currently done. Family studies take advantage of the fact that the same genetic variations will be more common in families — and thus easier to detect. However, the disadvantage of family studies is that the detected genetic variations may not be widespread in the population.
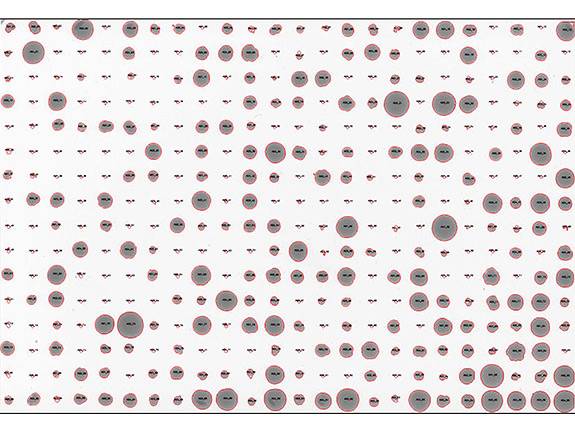
The researchers placed the more than 1,000 single-cell yeast offspring in a grid pattern on plates such as this one, and subjected each plate to a different condition, including different temperatures, food sources and presence of various drugs. Offspring thrived (large dots) or did not thrive (small dots) depending on their genetic make-up. Researchers examined the yeast to find out how much of their ability to survive was inherited from their parents. (Image courtesy of Joshua Bloom, Kruglyak Lab)
For the study in yeast, the team examined the offspring of two yeast cells, one that is commonly used in laboratory studies and the other in wine making. Although yeast usually reproduce asexually, under certain conditions, such as lack of food, two yeast cells will mate and produce offspring that, like human children, receive roughly half their genetic material from each parent. "Our study involves thousands of 'kids' from a single set of parents," Kruglyak said.
The team first sequenced the genomes of the two parent cells and then conducted scans for DNA variations in the genomes of 1,008 offspring. Yeast do not inherit height or disease risk from their parents, but they can inherit the ability to survive in adverse conditions. The researchers tested the parents and their offspring for the ability to grow under various conditions, including different temperatures, acidity levels, food sources, antibiotics, metal compounds, and in drugs such as caffeine.
The researchers then looked for associations between the DNA variations inherited from the parents and growth ability, and determined that the DNA variations accounted for nearly all of the resilience noted in the offspring.
The paper, "Finding the sources of missing heritability in a yeast cross," was published in Nature on Feb. 3, 2013. This work was supported by National Institutes of Health (NIH) grants R37 MH59520 and R01 GM102308; a James S. McDonnell Centennial Fellowship (L.K.); the Howard Hughes Medical Institute (L.K.); a National Science Foundation (NSF) fellowship (J.S.B.); an NIH postdoctoral fellowship F32 HG51762 (I.M.E.); and NIH grant P50 GM071508 to the Center for Quantitative Biology at the Lewis-Sigler Institute of Princeton University.