

The Landweber Lab

 |
The Landweber Lab
|
|
Research :
The current explosion of activity in comparative genomics and molecular
biology has permitted us to study the process of evolution at its most
fundamental level. DNA sequence analysis, for example, provides us
with insight into the mechanisms of selection and evolution at the level
of the gene. The discoveries of catalytic RNA, furthermore, and unusual forms of RNA splicing have led to
advances
in the study of the origin of life, and suggest that there are other
"molecular fossils," or primitive biological mechanisms,
still present
in modern species (Landweber 2007). Protists, in particular, have surprised
molecular biologists
with a bewildering
diversity of genome organization (e.g. Swart et al. 2013), from the impressive
scrambled genes in ciliates
to bizarre forms of RNA processing, including splicing and RNA editing,
as well as an abundance of nonstandard genetic codes. Therefore they
are a natural place to study primitive or aberrant genetic systems. 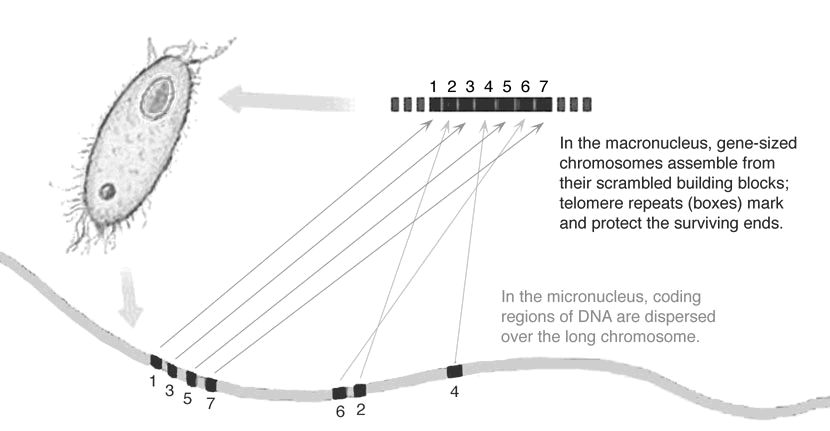
Current functional lines of experiments use RNAi or microinjection of small (27nt) or long noncoding RNA molecules to manipulate the pathway of genome rearrangement (see Fang et al. 2012, Cell and Nowacki et al. 2008, Nature, for two respective examples). Earlier work in our lab used test-tube evolution experiments to test the emergence of catalytic function from random RNA sequences (Landweber and Pokrovskaya PNAS 1999) and also to develop robust quantitative tests for the role of RNA-amino acid interactions in the origin of the Genetic Code (see Knight and Landweber 1998, 2000). Perhaps surprisingly, this same in vitro selection procedure also allowed us to construct a molecular computer out of RNA that can find and describe solutions to small mathematical search problems (Faulhammer et al. PNAS 2000). The ability to isolate new ribozymes from random sequences (e.g. Landweber and Pokrovskaya PNAS 1999) and to infer properties present in the last common ancestor of all life (Goldman et al. 2013) has fueled excitement about the possibility of discovering early pathways of RNA and protein evolution (e.g. Goldman et al. 2013). Ultimately, such surveys will make the world of possible primordial enzyme functions accessible even when the molecules are no longer present in modern species.
Full list of journal publications with links to abstracts in
medline: 
 9
Free open-access reprints available from PNAS here.
9
Free open-access reprints available from PNAS here.
Books
|