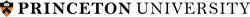Research
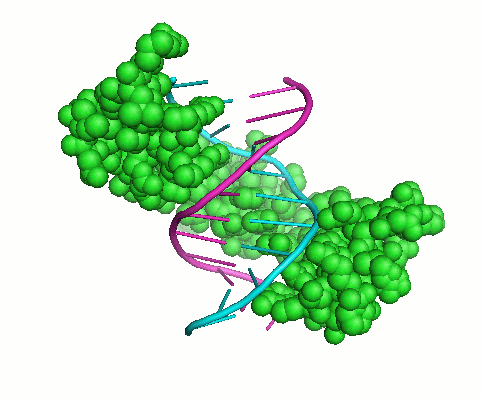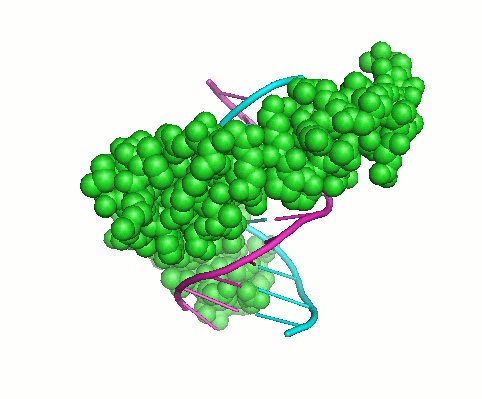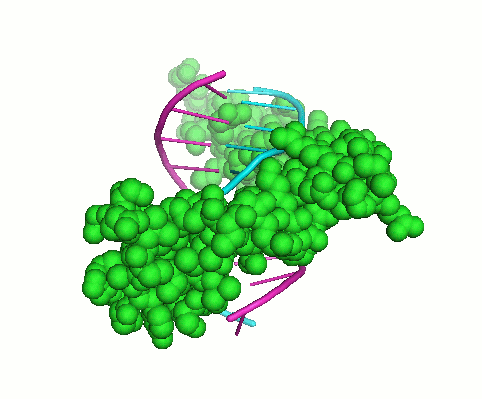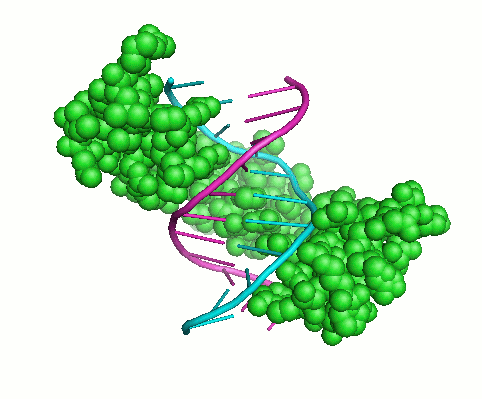
My overall research interests are to understand sequence-structure-function relationships in proteins and the specificity of macromolecular interactions. Interactions of proteins with other proteins and polynucleotides underlie all biological functions. These interactions establish the architecture of the cell and extracellular matrix, regulate biosynthesis, play a key role in signaling, underlie the assembly of macromolecular machines and mediate chemical transformations. Proteins are frequently constructed in a modular fashion from a limited number of more or less common protein domains. These domains mediate many protein functions: they can mediate protein-protein or protein-DNA interactions, target proteins to a specific subcellular location, nucleate the formation of multiprotein complexes, and control the conformation, activity, and substrate specificity of enzymes. Although we now have fairly complete list of the protein domains found in various organisms, our knowledge of how and why proteins interact with other molecules is limited. Knowledge of the basic principles of structure and function of protein domains, the ability to determine the binding specificity of these domains and to predict their physiological interacting partners are key steps in understanding many biological processes and molecular mechanisms of many human disorders.My work has been focused on two problems that are important in this connection: (1) studying collagens and collagen-like motifs essential for adopting triple-helical structure and binding ligands; and (2) understanding how the interaction specificity is encoded in protein sequences and structures, including the Cys2His2 zinc finger and the SH3 binding motifs. Both computational and experimental methods are needed to accelerate discovery and understanding in these areas. Experimental approaches include peptide models, binding assays, mutation analysis, differential scanning calorimetry, isothermal titration calorimetry, and a wide range of spectroscopic techniques. Computational approaches include structure-based modeling, crystal structure analysis, molecular dynamic simulations, sequence analysis techniques and machine learning. I always try to integrate both approaches, tackling the complex problems of characterizing, analyzing, designing and predicting protein interaction specificity by studying domains with modular structures. In general my goals are: (1) to develop and apply techniques to assess the interaction specificity of biologically interesting protein families in vitro, (2) to achieve an understanding of how specificity is encoded biophysically, through the analysis of protein sequence and structure, and (3) to develop and test computational methods for predicting and designing specific protein interactions.