When humans see their children about to eat something they oughtn't, we can simply tell them, "Don't eat that. It'll make you sick." Those who listen to this advice are spared the painful experience of learning that lesson for themselves. While other animals can't sit their offspring down for a good talking-to, that doesn't mean they are unable to instruct their descendants about potential harms.
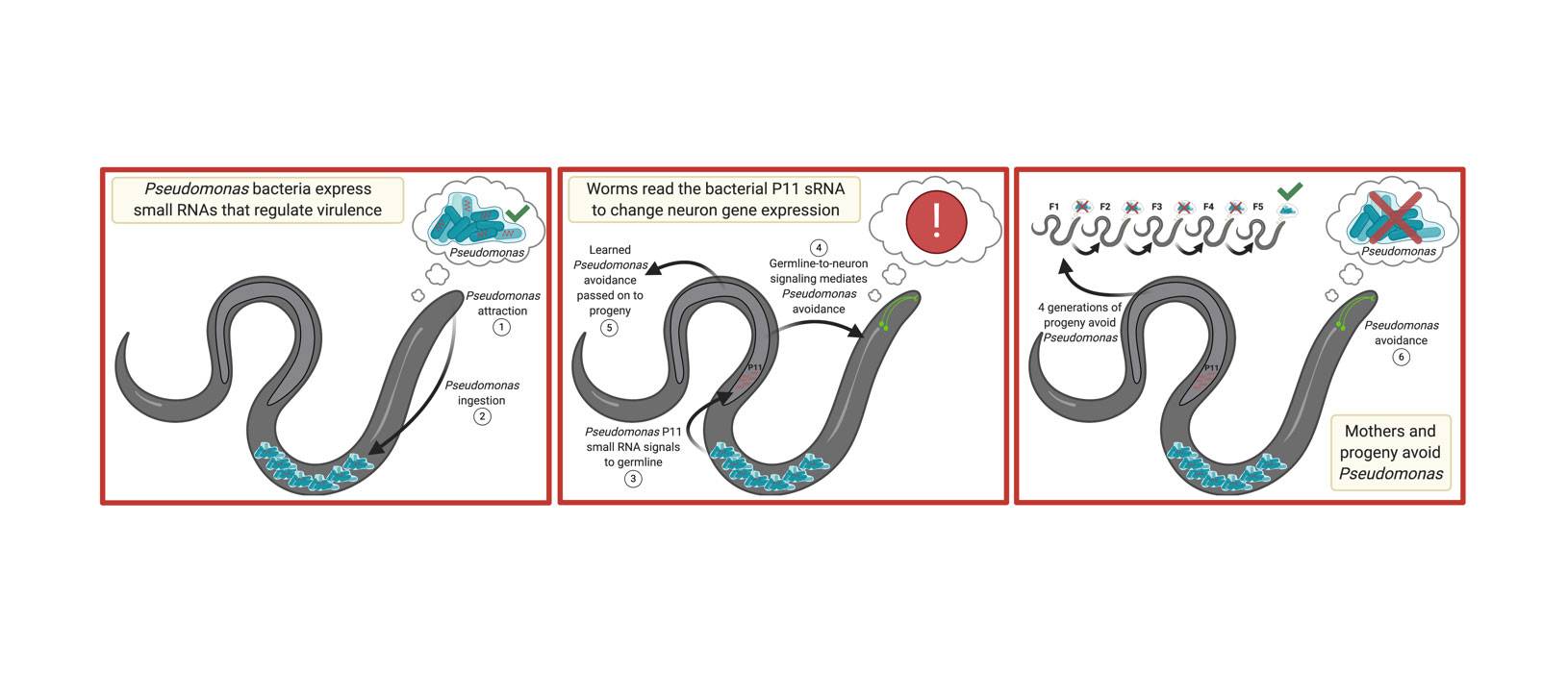
For instance, the microscopic roundworm Caenorhabditis elegans feeds on bacteria such as Pseudomonas aeruginosa. However, certain environmental conditions can cause P. aeruginosa to change in such a way that it becomes pathogenic -- that is, it sickens worms that eat it. In 2019, researchers in the Murphy lab showed that when worm mothers are made ill by P. aeruginosa, they learn to avoid the bacterium. Not only that, but their offspring, all the way down to their great-great grandchildren, somehow also know to avoid the bacterium. After four generations, though, the transgenerational avoidance behavior disappears, letting the worms return to feeding on the bacterium once more.
C. elegans has been so intensively studied that we not only know the identity (e.g., neuron, muscle cell, intestinal cell, etc.) of every cell in its body, but also the order in which it appears during embryonic development. Nonetheless, the worm still manages to surprise us with complex behaviors such as transgenerational avoidance. What causes this behavior? Initial work by the Murphy lab showed that other types of pathogenic bacteria didn't cause avoidance behavior in offspring, indicating the behavior is specific to pathogenic P. aeruginosa. Furthermore, the researchers showed that avoidance of pathogenic P. aeruginosa is controlled by one particular worm neuron. But several questions about the phenomenon remained, so the team continued to investigate.
"We wondered how the worms can know the identity of the bacteria they are eating," said Coleen Murphy, a professor of molecular biology and the Lewis-Sigler Institute for Integrative Genomics (LSI) and the senior author of a paper detailing the team's findings in the journal Nature.
To investigate this question, two Murphy lab researchers -- associate research scholar Rachel Kaletsky and graduate student Rebecca Moore -- fed worms with a harmless bacterium that had been spiked with different materials isolated from pathogenic P. aeruginosa. These materials included substances involved in the bacterium's metabolic processes, as well as bacterial genetic material. The latter is composed of three classes of molecules: DNA, which can be thought of as a cookbook containing recipes for all the proteins the organism needs to make to survive; messenger RNAs, which are copies of individual recipes that are read out to build proteins; and small RNAs, which don't code for proteins but instead perform regulatory functions in cells, often by promoting the destruction of messenger RNAs.
"Instead of a signal from the metabolites that the bacteria produce, as we had originally expected, we found that the worms 'read' the small RNAs that bacteria make, particularly small RNAs that correlate with the pathogenic state of the bacteria," said Murphy. In fact, the researchers discovered that inherited avoidance behavior requires one specific bacterial small RNA, called P11.
"The bacterial work that Geoff Vrla in Zemer Gitai's lab did was critical for proving that the key small RNA was P11," Murphy said. Gitai is the Edwin Grant Conklin Professor of Biology at Princeton.
"The P11 small RNA itself doesn't even make the worms sick -- just detecting the presence of P11 is enough to make the worms avoid the bacteria, and to pass it on to four generations of progeny," Murphy added.
Kaletsky, Moore and their colleagues found that once a worm has eaten the bacterium, P11 is absorbed and processed by the worm intestine, then it finds its way into the worm's eggs and sperm. (C. elegans mothers are hermaphrodites, so they possess both types of reproductive cells and can self-fertilize.) From there, P11 is conveyed to the neuron that controls avoidant behavior in the mother, where it causes destruction of worm messenger RNAs that encode a protein called macoilin. Loss of the messenger RNAs prevents manufacture of this protein, and this in turn causes the worm to avoid the pathogenic bacterium. P11's presence in the worm's reproductive cells also ensures it is available to subsequent generations of worms.
"As far as we know, this is the first example found of an animal host 'reading' the small RNA of a pathogen and evolving a response that helps it stay healthier, a kind of nascent adaptive immune system response," Murphy said.
"There are only a few examples of such inter-species molecular communication via small RNAs, and even fewer examples of adaptive transgenerational behavioral changes in response to small RNAs," said Julie Claycomb, the Canada Research Chair in Small RNA Biology at the University of Toronto, who was not involved in the work.
"This study sets a high standard for understanding the molecular mechanisms governing such phenomena, and opens a novel area of investigation going forward," added Claycomb.
"C. elegans interprets bacterial non-coding RNAs to learn pathogenic avoidance," by Rachel Kaletsky, Rebecca S. Moore, Geoffrey D. Vrla, Lance R. Parsons, Zemer Gitai and Coleen T. Murphy, was published in the Sept. 9 issue of the journal Nature (DOI: 10.1038/s41586-020-2699-5). This work was supported by a Pioneer Award (NIGMS DP1GM119167), the Glenn Foundation for Medical Research (GMFR CNV1001899), and the HHMI-Simons Faculty Scholar Program (AWD1005048) to C.T.M.; R.S.M. and G.D.V. were supported by T32GM007388 (NIGMS), and Z.G. by a Pioneer Award (DP1A1124669).