Effort could lead to improved, targeted therapies
A team of Princeton University scientists has produced a systematic listing of the ways a particular cancerous cell has "gone wrong," giving researchers a powerful tool that eventually could make possible new, more targeted therapies for patients.
"For a very long time, cancer therapies have been developed by trial and error to essentially kill a broad variety of rapidly dividing cells, good and bad -- that's why they have massive side effects," said Saeed Tavazoie, a professor in the Department of Molecular Biology and the Lewis-Sigler Institute for Integrative Genomics, who led the research. "The goal of cancer biology is to come up with therapies that are much more rational in terms of attacking the pathways that have been co-opted by cancer cells. The big challenge is to discover these pathways so that we can restore them to their normal state."
Writing in the Dec. 11 issue of Molecular Cell, Tavazoie, along with his colleagues Hani Goodarzi, a graduate student in molecular biology, and Olivier Elemento, a former postdoctoral researcher in the department, found they were able to systematically categorize and pinpoint the alterations in cancer pathways and to reveal the underlying regulatory code in DNA. Elemento is now on the faculty of Weill Cornell Medical College in New York.
"We are discovering that there are many components inside the cell that can get mutated and give rise to cancer," Tavazoie said. "Future cancer therapies have to take into account these specific pathways that have been mutated in individual cancers and treat patients specifically for that."
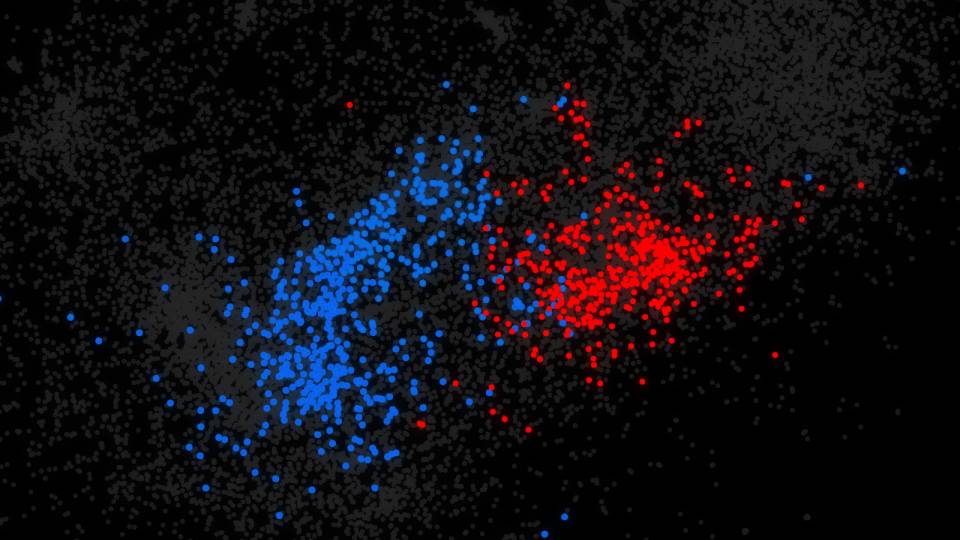
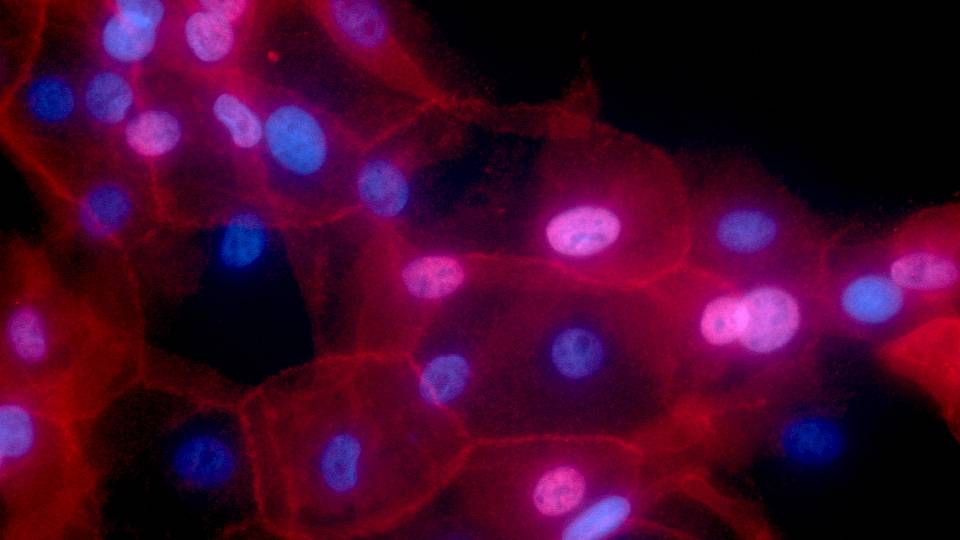
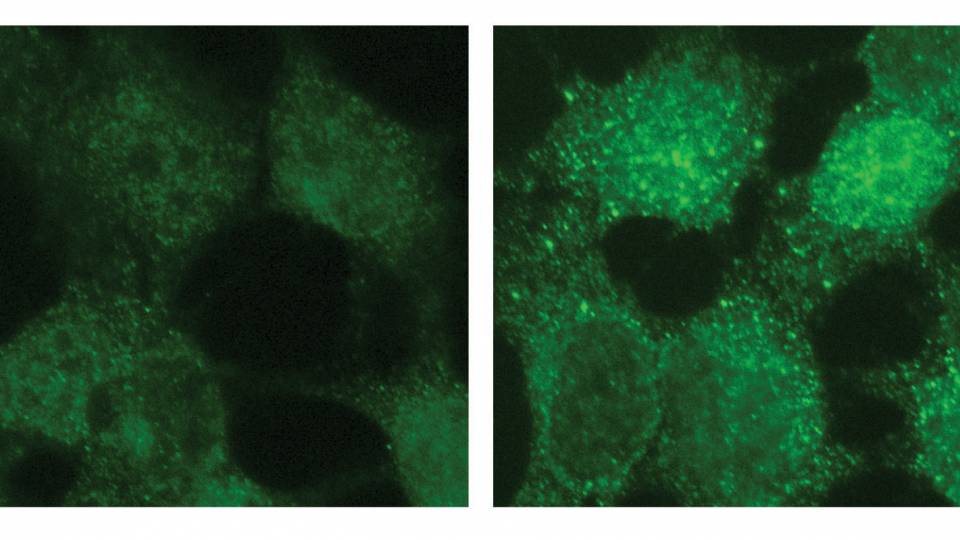
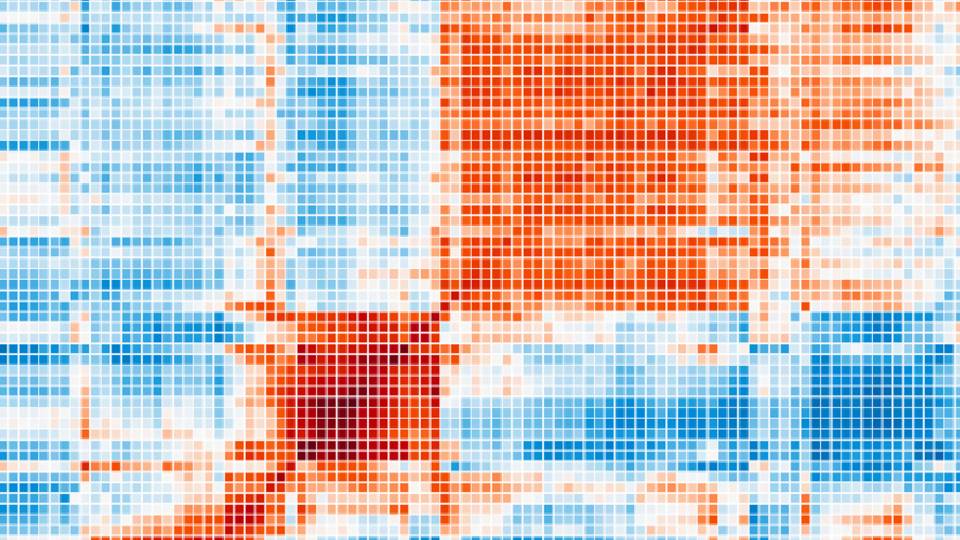
The researchers developed an algorithm, a problem-solving computer program that sorts through the behavior of each of 20,000 genes operating in a tumor cell. When genes are turned "on," they activate or "express" proteins that serve as signals, creating different pathways of action. Cancer cells often act in aberrant ways, and the algorithm can detect these subtle changes and track all of them.
"At the present moment, we lump a lot of cancers together and use the same therapy," Tavazoie said. "In the future, we are aiming to be much more precise about treating the exact processes that were perturbed by the mutations."
Pathologists presently examining the tumors of sick patients analyze a small set of tumor characteristics in order to determine the diagnostic and prognostic class to which the cells belong. This new method could give practitioners an encyclopedic accounting of the alterations in problem cells, spelling out the nature of the disease in much greater detail.
The algorithm devised by the group scans the DNA sequence of a given cell -- its genome -- and deciphers which sequences are controlling what pathways and whether any are acting differently from the norm. By deciphering the patterns, the scientists can conjure up the genetic regulatory code that is underlying a particular cancer.
The scientists developed the technique by employing modern methods of systems biology, where researchers seek to understand how components of living systems like cells work together to orchestrate processes, using powerful computers to sort vast arrays of data.
"Part of the promise of genomics and systems biology is the discovery of specific pathways of disease and finding ways to target them precisely," Tavazoie said. "We have focused on revealing what these pathways are."
The challenge for others, he said, will be to design specific therapies for such diseases, a process that could take many years. "This is an important first step," Tavazoie added.
The method ultimately could work for any type of cancer and paves the way for rational approaches to treating a host of other diseases from diabetes to neurological disorders, the scientists said.
The research was funded by the National Human Genome Research Institute of the National Institutes of Health.