Clifford Brangwynne (left), assistant professor of chemical and biological engineering, worked with postdoctoral researcher Ming-Tzo Wei and interdisciplinary collaborators to develop a method for looking at watery structures inside cells.
A research team led by Princeton engineers has revealed in remarkable new detail how liquid droplets can develop structure amidst the soup of material found inside a living cell. These droplets, known as membraneless organelles, play critical roles in cellular function and diseases.
The team, a mix of biologists and materials scientists, has shown that surprisingly low concentrations of proteins can readily condense into a droplet that has internal structure, yet is very dilute, consisting mostly of empty, watery space. This liquid scaffolding lets molecules only of certain sizes easily diffuse in and out of the structure, enabling them to perform their vital tasks.
The new insights into the molecular organization inside membraneless organelles will help clarify their contributions to health and — when that organization breaks down — to certain diseases.
“In this study, we have measured important aspects of the protein-to-protein interactions that drive the form and function of a membraneless organelle,” said Ming-Tzo Wei, a postdoctoral research associate in the Department of Chemical and Biological Engineering and lead author of the study published June 26 in the journal Nature Chemistry.
“We’re really starting to understand the molecular-level organization within this membraneless class of cellular structures,” said Clifford Brangwynne, an assistant professor of chemical and biological engineering, senior author of the paper and principal investigator of the Soft Living Matter Group.
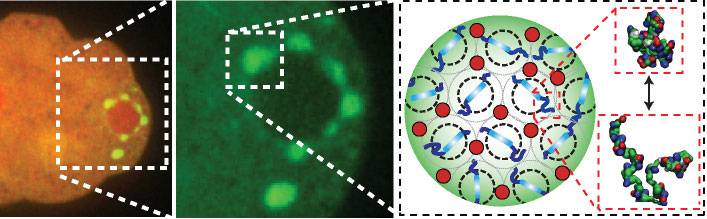
At left: Membraneless organelles, called P granules, are shown in green around a cell's nucleus in a flatworm embryo. Middle: A zoom-in of the liquid-like organelles. At right: An artist's impression of a tighter zoom into the P granule, revealing its structure that it is permeable to molecules only of certain sizes, shown in red.
The team collaborated with Rohit Pappu, a biomedical engineer at Washington University in St. Louis, and also included Rodney Priestley, associate professor of chemical and biological engineering, and Craig Arnold, director of the Princeton Institute for the Science and Technology of Materials.
The researchers focused on a protein type, LAF-1, that joins with other proteins and RNA to form a globular, membraneless organelle called a P granule. In a popularly studied roundworm, Caenorhabditis elegans, the P granules keep the worm’s sex cells in a prepared state for reproduction.
A set of experiments sought to determine the concentration of LAF-1 inside of a P granule versus the levels of the protein otherwise floating freely within the cell. Knowing the difference would tell the researchers what concentration of the protein is needed to form the structure. A novel technique, called ultrafast-scanning fluorescence correlation spectroscopy, proved critical to the task.
Developed in collaboration with paper co-author Arnold, who also is a professor of mechanical and aerospace engineering, the technique uses a special lens to reduce uncertainty about the size of a volume being scanned by a microscope. As a result, the concentration of proteins fitted with fluorescent tags can be accurately determined in a given space, for instance within a P granule.
Wei took a series of such measurements, along with co-first author Shana Elbaum-Garfinkle, also a postdoctoral research associate in the Department of Chemical and Biological Engineering. In addition, the researchers tracked the motions of molecules in the P granule and observed how interactions with RNA reduced the protein concentration, in effect lowering the granule’s fluid consistency, or viscosity.
For further insight, the researchers turned to the science of polymers, which are substances composed of many similar, smaller units, like those found in consumer plastic products. LAF-1 is a “disordered” protein, and can be thought of as a flexible polymer chain. The polymeric nature of LAF-1 allows it to form a scaffold-like network within the droplet. However, unlike with plastics, the team’s results indicated that the “mesh size,” or average size of the gaps between units, is relatively large, three to eight nanometers (billionths of a meter). Molecules larger than this span cannot move throughout the droplet. This result places limits on the kinds of material that the membraneless organelle can interact with inside of a cell, shedding light on its function.
The findings were further validated by a series of computer simulations run by computational biophysicist and co-first author Alex Holehouse, a graduate student working closely with his adviser Pappu of Washington University in St. Louis.
“We were able to basically swim inside the organelles to determine how much room is actually available," Pappu said in a news story published by Washington University. "While we expected to see a crowded swimming pool, we found one with plenty of room, and water. We’re starting to realize that these droplets are not all going to be the same.”
Pappu added that the implications for the work are broad. “It is essential to be able to understand how one can regulate the functions of these droplets,” Pappu said. “If we succeed, the impact could be transformative: it’s not just cancer, it’s neurodegeneration, about developmental disorders, and even the fundamentals of cell biology.”
The advance required the melding of multiple perspectives and expertise, Brangwynne said.
“This study represents a unique collaboration between soft matter and polymer physics, mechanical engineering, computational physics and biology,” said Brangwynne. “Working together in this way has given us all a real sense of triumph in having helped move science forward.”
Additional authors on the paper include Carlos Chih-Hsiung Chen, a research specialist in the Department of Chemical and Biological Engineering, and Marina Feric, formerly of Princeton and now a postdoctoral fellow at the National Institutes of Health. The work was supported by the Princeton Center for Complex Materials, the National Science Foundation, the National Institutes of Health and the Eric and Wendy Schmidt Transformative Technology Fund.



