Proper embryonic development of the fruit fly Drosophila melanogaster is governed by patterns of protein activity bequeathed to the fertilized egg by its mother. While the embryo is still a single cell, the maternal cells surrounding it deposit certain proteins inside it at specific locations. This establishes protein gradients that direct the development of embryonic features along its anterior-posterior and ventral-dorsal axes. Later, the embryo receives another round of maternal information, called terminal patterning, that guides the development of its head and tail.
Terminal patterning is driven by a protein called Torso that is made by the mother and deposited throughout the embryo. Torso is stimulated by binding to other proteins that are also produced by the mother, but present only at the embryo’s anterior and posterior ends. Torso stimulation kicks off several signaling cascades, including one called the Ras/ERK pathway, whose activation directs the expression of genes crucial to embryonic development. Mothers lacking Torso or the proteins to which it binds are sterile because their embryos fail to develop head and tail structures and eventually die.
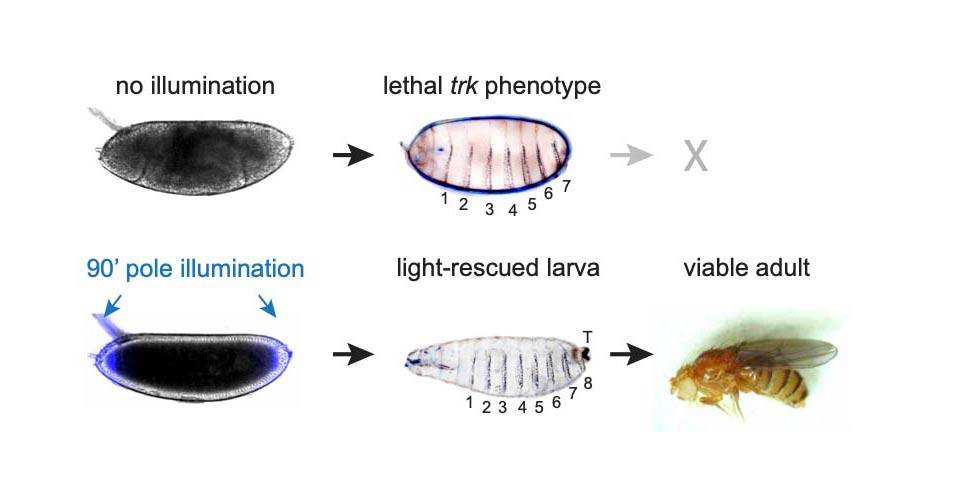
Princeton University researchers explored embryonic development using optogenetics, using light to stimulate modified fruit fly embryos. They found that while all of their modified embryos that were left in the dark died after failing to develop properly, about a third of the light-stimulated embryos developed normally.
“At this point, most developmental patterns have been studied in great detail, so biologists know when and where they appear and disappear,” said Jared Toettcher, an assistant professor of molecular biology. “What is unknown is exactly what information is contained in such a pattern.”
For instance, the level at which a patterning protein is expressed may convey information about identity or purpose (i.e., what genes should be expressed) to an embryonic cell. Alternatively, this information could be encoded by other factors, such as where or for how long the protein is expressed. Traditional approaches to studying the functions of a given protein cannot manipulate protein expression with enough spatial or temporal precision to address such questions. However, Toettcher’s lab had earlier used the emerging technology of “optogenetics” to develop a new research tool: a protein called OptoSOS that, when expressed in cells, allows researchers to activate the Ras/Erk signaling pathway while OptoSOS is illuminated with blue light.
“This is great because we can produce light patterns with high precision, allowing us to draw any pattern that we would like onto the embryo,” said Toettcher.
To investigate where and when Ras/Erk signaling is needed for terminal patterning, Johnson and his colleagues expressed OptoSOS in the embryos of flies that do not have Torso signaling. They then placed the embryos under a microscope and precisely stimulated the animals’ anterior and posterior ends with light for long enough to mimic the natural duration of Ras/Erk signaling. Next, they watched the embryos develop.
Remarkably, whereas all embryos left in the dark died after failing to develop properly, about a third of the light-stimulated embryos developed normally. Despite the trauma of the extraction procedure and of growing in a microscope chamber, these embryos formed head and tail structures, and eventually hatched and reached adulthood. Female flies created in this way even laid eggs—although, as expected, these eggs were sterile because like their mothers, the flies lacked Torso signaling, but unlike their mothers, their eggs were not stimulated with light.
Much of terminal patterning is driven by two genes whose expression is controlled by the Ras/Erk pathway in response to Torso signaling. In normal embryos, the expression of these two genes occurs in different cells at different times. In light-stimulated OptoSOS embryos lacking Torso signaling, however, expression of the two genes overlapped more in both space and time, suggesting their precise expression patterns are not required for development.
The authors next investigated whether different aspects of the developmental program are triggered at the same or different stimulus thresholds. To do this, they monitored embryonic development after varying the intensity and duration of light stimulus in OptoSOS embryos lacking Torso signaling.
“We found that the terminal pattern appears to work as a series of switches, where successively longer light pulses trigger a predictable sequence of body parts being ‘rescued’ one by one,” said Toettcher.
Together, these data suggest that for terminal signaling, what appears to be a very complex developmental program is actually under the control of a relatively simple system that depends on different thresholds of Ras/Erk signaling.
“These findings demonstrate the power of using optogenetics to activate and inactivate signaling pathways in time and space,” said Gertrude Schüpbach, a researcher whose work was instrumental in the early studies that identified the components of terminal patterning. “Even more amazingly, they are able to tune the strength of the signal and obtain precise quantitative information about the mechanism in the living embryo.”
“We think this is the beginning of mapping when and where patterns need to be delivered to produce specific body structures,” said Toettcher.
“Optogenetic rescue of a developmental patterning mutant” by Heath E. Johnson, Nareg J.V. Djabrayan, Stanislav Y. Shvartsman and Jared E. Toettcherwas published in Current Biology on July 23 (DOI: 10.1016/j.cub.2020.06.059). This work was supported by the NIH (Ruth Kirschstein F32GM119297, DP2EB024247 and 5R01HD085870) and a National Science Foundation CAREER Award (1750663).



